Wrobel Lab
Laboratory of Structural Cell Virology
© The Francis Crick Institute
Who we are
We are a small research group interested in molecular mechanisms underlying viral evolution and host changes. We are based at the Department of Biochemistry of the University of Oxford and we are grateful to receive funding from the Wellcome Trust and the ERC.
Below you will find the outline of our research, and recent publications. You can also use bookmarks above to find out more about our team, the opportunities to join us, and recent lab news.
Research Outline
Many deadly human pathogens, such as influenza and SARS viruses, are made up of relatively few components but can infect a number of different hosts. How is it possible that these components suffice to fulfil all the functions necessary for a virus to infect the cell and then to assemble into a new viral particle? How do viral proteins perform multiple functions and how does the virus manage to retain all these functions as it evolves? How does a virus infect different hosts using the same set of its own proteins to engage a range of machineries of different hosts? And, finally, how does a virus evolve and ‘learn’ to optimise its interactions with a new host?
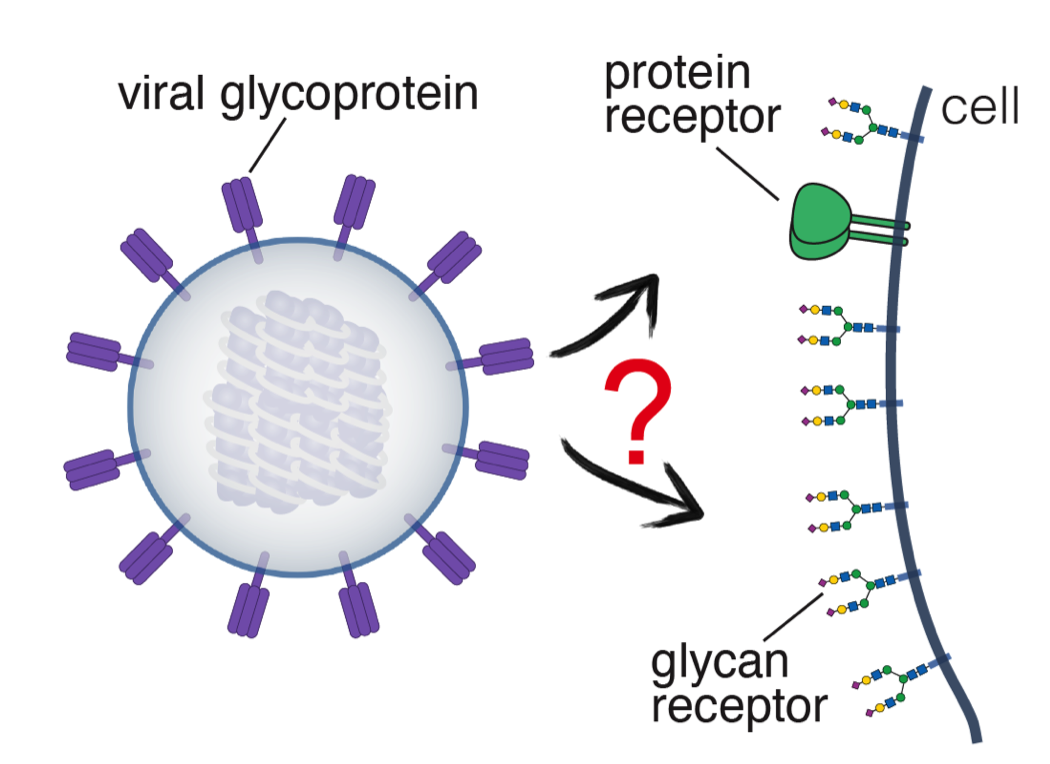
The recent COVID-19 pandemic has demonstrated that these questions are key to understanding where new viruses come from and how they evolve upon transmitting to a new host. We have used biochemical, biophysical, and structural methodologies, mainly cryoEM, to reveal the mechanisms by which SARS-CoV-2 became able to infect humans and then evolved further to optimise infectivity in the variants of concern. The lab continues to work on coronaviruses and studies influenza viruses to understand how their proteins achieve the versatility needed to infect diverse hosts and fulfil multiple functions during infection. In particular, we want to explain how related viral strains use similar glycoproteins to engage receptors as different as glycans and proteins. We are also interested in understanding how viruses assemble: how do viral components find one another in the dense environment of the cell, and how do cellular machineries facilitate or interfere with this process.
Tackling these questions can directly impact public health. The more we understand the rules governing the evolution of viral proteins, the better we can predict the impact of emerging viruses and thus increase our pandemic preparedness. Our long-term aim is to use structural and mechanistic insights to guide design of much-needed new antivirals and vaccines against zoonotic viruses.
Publications
#corresponding authors, *first authors
KEY PAPERS
1. Valeria Calvaresi#, Antoni G. Wrobel#, Joanna Toporowska, Dietmar Hammerschmid, Katie J. Doores, Richard T. Bradshaw, Ricardo B. Parsons, Donald J. Benton, Chloë Roustan, Eamonn Reading, Michael H. Malim, Steven J. Gamblin, Argyris Politis#. (2023) “Structural dynamics in the evolution of SARS-CoV-2 spike glycoprotein” Nature Communications, 14 (1)
2. Antoni G. Wrobel# (2023) “Mechanism and evolution of human ACE2 binding by SARS-CoV-2 spike” Current Opinions in Structural Biology 102619.
3. Antoni G. Wrobel*#, Donald J. Benton*#, Chloë Roustan, Annabel Borg, Saira Hussain, Stephen R. Martin, Peter B. Rosenthal, John J. Skehel, Steven J. Gamblin# (2022). “Evolution of the SARS-CoV-2 spike in the human host” Nature Communications 13, 1178.
4. Antoni G. Wrobel*#, Donald J. Benton*#, Pengqi Xu, Annabel Borg, Chloë Roustan, Stephen R. Martin, Peter B. Rosenthal, John J. Skehel, Steven J. Gamblin# (2021). “Structure and binding properties of Pangolin-CoV Spike glycoprotein inform the evolution of SARS-CoV-2.” Nature Communications, 12 (1), 837.
5. Donald J. Benton*#, Antoni G. Wrobel*#, Chloë Roustan, Annabel Borg, Pengqi Xu, Stephen R. Martin, Peter B. Rosenthal, John J. Skehel, Steven J. Gamblin#. (2021) “The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2” Proceedings of the National Academy of Sciences, 118(9), e2022586118.
6. Antoni G. Wrobel*#, Donald J. Benton*#, Saira Hussain, Ruth Harvey, Stephen R. Martin, Chloë Roustan, Peter B. Rosenthal, John J. Skehel, Steven J. Gamblin# (2020). “Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein” Nature Communications 11 (1), 5337.
7. Donald J. Benton*#, Antoni G. Wrobel*#, Pengqi Xu, Chloë Roustan, Stephen R. Martin, Peter B. Rosenthal, John J. Skehel, Steven J. Gamblin# (2020) “Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion” Nature 588(7837), 327–330.
8. Antoni G. Wrobel*#, Donald J. Benton*#, Pengqi Xu, Chloë Roustan, Stephen R. Martin, Peter B. Rosenthal, John J. Skehel, Steven J. Gamblin# (2020) “SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.” Nature Structural and Molecular Biology 27, 763–767 (2020).
RECENT PAPERS
1. Jefferson J.S. Santos, Shengyang Wang, Ryan McBride, Lorin Adams, Ruth Harvey, Yan Zhao, Antoni G. Wrobel, Steven Gamblin, John Skehel, Nicola S. Lewis, James C. Paulson, Scott E. Hensley (2025) “Bovine H5N1 binds poorly to human-type sialic acid receptors” Nature 640 E18–E20
2. Maria Kaukab Osman, Jonathan Robert, Lukas Broich, Dennis Frank, Robert Grosse, Martin Schwemmle, Antoni G. Wrobel, Kevin Ciminski, Christian Sieben, Peter Reuther (2025) “The bat Influenza A Virus subtype H18N11 induces nanoscale MHCII clustering upon host cell attachment” Nature Communications 16: 3847
3. Lewis Timimi, Antoni G. Wrobel, George N. Chiduza, Sarah L. Maslen, Antonio Torres-Méndez, Beatriz Montaner, Colin Davis, J. Mark Skehel, John L. Rubinstein, Anne Schreiber, Rupert Beale (2024) “The V-ATPase/ATG16L1 axis is controlled by the V1H subunit” Molecular Cell 84 (15), 2966-2983, e9
4. Jim Baggen, Maarten Jacquemyn, Leentje Persoons, Els Vanstreels, Valerie E Pye, Antoni G. Wrobel, Valeria Calvaresi, Stephen R Martin, Chloë Roustan, Nora B Cronin, Eamonn Reading, Hendrik Jan Thibaut, Thomas Vercruysse, Piet Maes, Frederik De Smet, Angie Yee, Toey Nivitchanyong, Marina Roell, Natalia Franco-Hernandez, Herve Rhinn, Alusha Andre Mamchak, Maxime Ah Young-Chapon, Eric Brown, Peter Cherepanov, Dirk Daelemans (2023) “TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry” Cell 186 (16), 3427-3442. e22
5. Edgar Gonzalez-Rodriguez, Mia Zol-Hanlon, Ganka Bineva-Todd, Andrea Marchesi, Mark Skehel, Keira Mahoney, Chloe Roustan, Annabel Borg, Lucia Di Vagno, Svend Kjaer, Antoni G. Wrobel, Donald J Benton, Philipp Nawrath, Sabine L Flitsch, Dhira Joshi, Andres Manuel Gonzalez-Ramirez, Katalin A Wilkinson, Robert J Wilkinson, Ramon Hurtado-Guerrero, Stacy A Malaker, Benjamin Schuman. (2023) “O-Linked Sialoglycans Modulate the Proteolysis of SARS-CoV-2 Spike and Contribute to the Mutational Trajectory in Variants of Concern” ACS Central Science 9 (3), 393-404
6. Okikiola M Olajide, Maria Kaukab Osman, Jonathan Robert, Susanne Kessler, Lina Kathrin Toews, Thiprampai Thamamongood, Jacques Neefjes, Antoni G. Wrobel, Martin Schwemmle, Kevin Ciminski, Peter Reuther (2023) “Evolutionarily conserved amino acids in MHC-II mediate bat influenza A virus entry into human cells” PLOS Biology 21 (7), e3002182
7. Lena Jaki, Sebastian Weigang, Lisa Kern, Stefanie Kramme, Antoni G. Wrobel, Andrea B Grawitz, Philipp Nawrath, Stephen R Martin, Theo Dähne, Julius Beer, Miriam Disch, Philipp Kolb, Lisa Gutbrod, Sandra Reuter, Klaus Warnatz, Martin Schwemmle, Steven J Gamblin, Elke Neumann-Haefelin, Daniel Schnepf, Thomas Welte, Georg Kochs, Daniela Huzly, Marcus Panning, Jonas Fuchs (2023) “Total escape of SARS-CoV-2 from dual monoclonal antibody therapy in an immunocompromised patient” Nature Communications 13, 1178
8. Thao T. Le, Donald J. Benton, Antoni G. Wrobel, Steven J. Gamblin. “Development of high affinity broadly reactive aptamers for spike protein of multiple SARS-CoV-2 variants” (2023) RSC advances 13 (22), 15322-15326
9. Philipp Nawrath, Antoni G. Wrobel# (2022). “Hold your horses: The receptor-binding domains of SARS-CoV-2, SARS-CoV, and hCoV-NL63 bind equine ACE2” Structure 30, 1367-1368. (preview)
10. Nathan Zaccai, Zuzana Kadlecova, Veronica Kane Dickson, Kseniya Korobchevskaya, Jan Kamenicky, Oleksiy Kovtun, Perunthottathu K Umasankar, Antoni G Wrobel, Jonathan GG Kaufman, Sally R Gray, Kun Qu, Philip R Evans, Marco Fritzsche, Filip Sroubek, Stefan Höning, John AG Briggs, Bernard T Kelly, David J. Owen; Linton M. Traub (2022). “FCHO controls AP2’s initiating role in endocytosis through a PtdIns(4,5)P2-dependent switch” Science Advances 22 (8), eabn2018
11. Kevin W. Ng*, Nikhil Faulkner*, Antoni G. Wrobel, Steve Gamblin, George Kassiotis (2021) “Heterologous humoral immunity to human and zoonotic coronaviruses: aiming for the Achilles heel” Seminars in Immunology, 55, 101507
12. Annachiara Rosa, Valerie E Pye, Carl Graham, Luke Muir, Jeffrey Seow, Kevin W Ng, Nicola J Cook, Chloe Rees-Spear, Eleanor Parker, Mariana Silva Dos Santos, Carolina Rosadas, Alberto Susana, Hefin Rhys, Andrea Nans, Laura Masino, Chloe Roustan, Evangelos Christodoulou, Rachel Ulferts, Antoni G Wrobel, Charlotte-Eve Short, Michael Fertleman, Rogier Sanders, Judith Heaney, Moira Spyer, Svend Kjær, Andy Riddell, Michael H Malim, Rupert Beale, James MacRae, Graham Taylor, Eleni Nastouli, Marit J van Gils, Peter B Rosenthal, Massimo Pizzato, Myra O McClure, Richard S Tedder, George Kassiotis, Laura E McCoy, Katie J Doores, Peter Cherepanov (2021). “SARS-CoV-2 can recruit a haem metabolite to evade antibody immunity”. Science Advances, 22 (7), eabg7607
13. Kevin W Ng, Nikhil Faulkner, Georgina H Cornish, Annachiara Rosa, Ruth Harvey, Saira Hussain, Rachel Ulferts, Christopher Earl, Antoni G Wrobel, Donald J Benton, Chloe Roustan, William Bolland, Rachael Thompson, Ana Agua-Doce, Philip Hobson, Judith Heaney, Hannah Rickman, Stavroula Paraskevopoulou, Catherine F Houlihan, Kirsty Thomson, Emilie Sanchez, Gee Yen Shin, Moira J Spyer, Dhira Joshi, Nicola O’Reilly, Philip A Walker, Svend Kjaer, Andrew Riddell, Catherine Moore, Bethany R Jebson, Meredyth Wilkinson, Lucy R Marshall, Elizabeth C Rosser, Anna Radziszewska, Hannah Peckham, Coziana Ciurtin, Lucy R Wedderburn, Rupert Beale, Charles Swanton, Sonia Gandhi, Brigitta Stockinger, John McCauley, Steve J Gamblin, Laura E McCoy, Peter Cherepanov, Eleni Nastouli, George Kassiotis (2020). “Pre-existing and de novo humoral immunity to SARS-CoV-2 in humans” Science, 370(6522), 1339–1343.
14. Kevin W. Ng, Jan Attig, William Bolland, George R. Young, Jack Major, Antoni G. Wrobel, Steve Gamblin, Andreas Wack, George Kassiotis (2020). “Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option” Nature Genetics, 52(12), 1294–1302
© The Francis Crick Institute